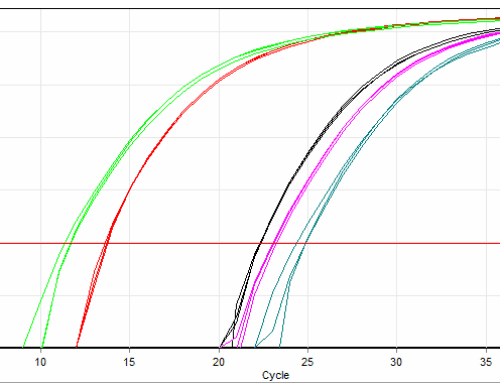
qPCR validation of next generation sequencing results
qPCR validation of next-generation sequencing (NGS) results remains critical for ensuring data accuracy, resolving technical limitations, and meeting publication or clinical standards. While NGS enables high-throughput genomic analysis, qPCR provides targeted verification through its simplicity, sensitivity, and cost-effectiveness, particularly for low-abundance targets or clinically actionable variants.
Key Reasons for qPCR Validation
- Error Mitigation:
NGS workflows involve complex sample preparation, bioinformatics pipelines, and potential biases (e.g., GC-rich region underrepresentation)[1]. qPCR reduces false positives/negatives by independently confirming variants or expression levels[2][3]. - Sensitivity for Low-Abundance Targets:
qPCR outperforms NGS in detecting low-copy transcripts or rare variants (<1% allelic frequency) due to its linear amplification and lack of reliance on sequencing depth[1][3]. - Reference Genome Limitations:
NGS aligns reads to error-containing reference genomes, which may disregard SNPs or structural variations. qPCR uses actual templates, bypassing alignment biases[1].
Validation Workflow Integration
- Orthogonal Verification:
qPCR validates NGS-identified variants or differentially expressed genes. For example, COVID-19 variant tracking combined NGS with RT-qPCR assays targeting spike protein deletions, achieving >95% concordance[4].
| Method | Role in NGS Validation | Key Advantage |
| qPCR | Target-specific verification | Cost-effective, high-throughput |
Best Practices
- Assay Design:
Use TaqMan probes or FRET-based assays with in silico/in vitro validation to ensure primer specificity[4][5]. - Data Normalization:
Address NGS assumptions (e.g., uniform mRNA levels across samples) by validating housekeeping genes via qPCR[1][6]. - Guideline Compliance:
Follow Association of Molecular Pathology (AMP) recommendations for analytical validation, including reference materials and error-based quality controls[6].
Complementary Use Cases
- Upstream QC: Verify cDNA integrity pre-NGS using qPCR[2].
- Downstream Prioritization: Narrow NGS-derived gene lists to clinically relevant targets for high-confidence validation[3][5].
While emerging technologies like ddPCR enhance precision, qPCR remains indispensable for balancing NGS scalability with rigorous validation, particularly in clinical diagnostics and peer-reviewed research[1][6][2].
⁂
- https://sciencellonline.com/blog/value-of-qpcr-in-the-next-generation-sequencing-era/
- https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/ngs-comparison.html
- https://www.illumina.com/science/technology/next-generation-sequencing/beginners/advantages/ngs-vs-qpcr.html
- https://pmc.ncbi.nlm.nih.gov/articles/PMC10059866/
- https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-applications/gene-expression-using-real-time-pcr/verification-ngs-microarray-results.html
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6941185/